
AC T25732
XX
ID T25732
XX
DT 23.08.2006 (created); man.
DT 12.07.2013 (updated); ili.
CO Copyright (C), QIAGEN.
XX
FA Notch1-isoform1
XX
SY MOTCH A; MT14; neurogenic locus Notch homolog protein 1; NOTCH 1; Notch1 300kDa protein; Notch1 p300; Notch1 precursor; P300.
XX
OS mouse, Mus musculus
OC eukaryota; animalia; metazoa; chordata; vertebrata; tetrapoda; mammalia; eutheria; rodentia; myomorpha; muridae; murinae
XX
GE G006477 Notch1.
XX
SZ 2531 AA; 270.8 kDa (cDNA) (calc.).
XX
SQ MPRLLTPLLCLTLLPALAARGLRCSQPSGTCLNGGRCEVANGTEACVCSGAFVGQRCQDS
SQ NPCLSTPCKNAGTCHVVDHGGTVDYACSCPLGFSGPLCLTPLDNACLANPCRNGGTCDLL
SQ TLTEYKCRCPPGWSGKSCQQADPCASNPCANGGQCLPFESSYICRCPPGFHGPTCRQDVN
SQ ECSQNPGLCRHGGTCHNEIGSYRCACRATHTGPHCELPYVPCSPSPCQNGGTCRPTGDTT
SQ HECACLPGFAGQNCEENVDDCPGNNCKNGGACVDGVNTYNCRCPPEWTGQYCTEDVDECQ
SQ LMPNACQNGGTCHNTHGGYNCVCVNGWTGEDCSENIDDCASAACFQGATCHDRVASFYCE
SQ CPHGRTGLLCHLNDACISNPCNEGSNCDTNPVNGKAICTCPSGYTGPACSQDVDECALGA
SQ NPCEHAGKCLNTLGSFECQCLQGYTGPRCEIDVNECISNPCQNDATCLDQIGEFQCICMP
SQ GYEGVYCEINTDECASSPCLHNGHCMDKINEFQCQCPKGFNGHLCQYDVDECASTPCKNG
SQ AKCLDGPNTYTCVCTEGYTGTHCEVDIDECDPDPCHYGSCKDGVATFTCLCQPGYTGHHC
SQ ETNINECHSQPCRHGGTCQDRDNSYLCLCLKGTTGPNCEINLDDCASNPCDSGTCLDKID
SQ GYECACEPGYTGSMCNVNIDECAGSPCHNGGTCEDGIAGFTCRCPEGYHDPTCLSEVNEC
SQ NSNPCIHGACRDGLNGYKCDCAPGWSGTNCDINNNECESNPCVNGGTCKDMTSGYVCTCR
SQ EGFSGPNCQTNINECASNPCLNQGTCIDDVAGYKCNCPLPYTGATCEVVLAPCATSPCKN
SQ SGVCKESEDYESFSCVCPTGWQGQTCEVDINECVKSPCRHGASCQNTNGSYRCLCQAGYT
SQ GRNCESDIDDCRPNPCHNGGSCTDGINTAFCDCLPGFQGAFCEEDINECASNPCQNGANC
SQ TDCVDSYTCTCPVGFNGIHCENNTPDCTESSCFNGGTCVDGINSFTCLCPPGFTGSYCQY
SQ DVNECDSRPCLHGGTCQDSYGTYKCTCPQGYTGLNCQNLVRWCDSAPCKNGGRCWQTNTQ
SQ YHCECRSGWTGVNCDVLSVSCEVAAQKRGIDVTLLCQHGGLCVDEGDKHYCHCQAGYTGS
SQ YCEDEVDECSPNPCQNGATCTDYLGGFSCKCVAGYHGSNCSEEINECLSQPCQNGGTCID
SQ LTNSYKCSCPRGTQGVHCEINVDDCHPPLDPASRSPKCFNNGTCVDQVGGYTCTCPPGFV
SQ GERCEGDVNECLSNPCDPRGTQNCVQRVNDFHCECRAGHTGRRCESVINGCRGKPCKNGG
SQ VCAVASNTARGFICRCPAGFEGATCENDARTCGSLRCLNGGTCISGPRSPTCLCLGSFTG
SQ PECQFPASSPCVGSNPCYNQGTCEPTSENPFYRCLCPAKFNGLLCHILDYSFTGGAGRDI
SQ PPPQIEEACELPECQVDAGNKVCNLQCNNHACGWDGGDCSLNFNDPWKNCTQSLQCWKYF
SQ SDGHCDSQCNSAGCLFDGFDCQLTEGQCNPLYDQYCKDHFSDGHCDQGCNSAECEWDGLD
SQ CAEHVPERLAAGTLVLVVLLPPDQLRNNSFHFLRELSHVLHTNVVFKRDAQGQQMIFPYY
SQ GHEEELRKHPIKRSTVGWATSSLLPGTSGGRQRRELDPMDIRGSIVYLEIDNRQCVQSSS
SQ QCFQSATDVAAFLGALASLGSLNIPYKIEAVKSEPVEPPLPSQLHLMYVAAAAFVLLFFV
SQ GCGVLLSRKRRRQHGQLWFPEGFKVSEASKKKRREPLGEDSVGLKPLKNASDGALMDDNQ
SQ NEWGDEDLETKKFRFEEPVVLPDLSDQTDHRQWTQQHLDAADLRMSAMAPTPPQGEVDAD
SQ CMDVNVRGPDGFTPLMIASCSGGGLETGNSEEEEDAPAVISDFIYQGASLHNQTDRTGET
SQ ALHLAARYSRSDAAKRLLEASADANIQDNMGRTPLHAAVSADAQGVFQILLRNRATDLDA
SQ RMHDGTTPLILAARLAVEGMLEDLINSHADVNAVDDLGKSALHWAAAVNNVDAAVVLLKN
SQ GANKDMQNNKEETPLFLAAREGSYETAKVLLDHFANRDITDHMDRLPRDIAQERMHHDIV
SQ RLLDEYNLVRSPQLHGTALGGTPTLSPTLCSPNGYLGNLKSATQGKKARKPSTKGLACGS
SQ KEAKDLKARRKKSQDGKGCLLDSSSMLSPVDSLESPHGYLSDVASPPLLPSPFQQSPSMP
SQ LSHLPGMPDTHLGISHLNVAAKPEMAALAGGSRLAFEPPPPRLSHLPVASSASTVLSTNG
SQ TGAMNFTVGAPASLNGQCEWLPRLQNGMVPSQYNPLRPGVTPGTLSTQAAGLQHSMMGPL
SQ HSSLSTNTLSPIIYQGLPNTRLATQPHLVQTQQVQPQNLQLQPQNLQPPSQPHLSVSSAA
SQ NGHLGRSFLSGEPSQADVQPLGPSSLPVHTILPQESQALPTSLPSSMVPPMTTTQFLTPP
SQ SQHSYSSSPVDNTPSHQLQVPEHPFLTPSPESPDQWSSSSPHSNISDWSEGISSPPTTMP
SQ SQITHIPEAFK
XX
SC Swiss-Prot#Q01705-1
XX
FT 20 58  PS50026; EGF_3.
FT 23 58
PS50026; EGF_3.
FT 23 58  SM00181; EGF.
FT 59 99
SM00181; EGF.
FT 59 99  PS50026; EGF_3.
FT 59 99
PS50026; EGF_3.
FT 59 99  SM00179; EGF_CA.
FT 62 99
SM00179; EGF_CA.
FT 62 99  SM00181; EGF.
FT 63 98
SM00181; EGF.
FT 63 98  PF00008; EGF.
FT 102 139
PF00008; EGF.
FT 102 139  PS50026; EGF_3.
FT 105 139
PS50026; EGF_3.
FT 105 139  SM00181; EGF.
FT 106 138
SM00181; EGF.
FT 106 138  PF00008; EGF.
FT 106 139
PF00008; EGF.
FT 106 139  SM00179; EGF_CA.
FT 140 176
SM00179; EGF_CA.
FT 140 176  PS50026; EGF_3.
FT 140 176
PS50026; EGF_3.
FT 140 176  SM00179; EGF_CA.
FT 143 176
SM00179; EGF_CA.
FT 143 176  SM00181; EGF.
FT 144 175
SM00181; EGF.
FT 144 175  PF00008; EGF.
FT 178 215
PF00008; EGF.
FT 178 215  PF07645; EGF_CA.
FT 178 216
PF07645; EGF_CA.
FT 178 216  PS50026; EGF_3.
FT 178 216
PS50026; EGF_3.
FT 178 216  SM00179; EGF_CA.
FT 181 216
SM00179; EGF_CA.
FT 181 216  SM00181; EGF.
FT 218 255
SM00181; EGF.
FT 218 255  PS50026; EGF_3.
FT 221 255
PS50026; EGF_3.
FT 221 255  SM00181; EGF.
FT 222 254
SM00181; EGF.
FT 222 254  PF00008; EGF.
FT 222 255
PF00008; EGF.
FT 222 255  SM00179; EGF_CA.
FT 257 293
SM00179; EGF_CA.
FT 257 293  PS50026; EGF_3.
FT 257 293
PS50026; EGF_3.
FT 257 293  SM00179; EGF_CA.
FT 260 293
SM00179; EGF_CA.
FT 260 293  SM00181; EGF.
FT 261 292
SM00181; EGF.
FT 261 292  PF00008; EGF.
FT 295 332
PF00008; EGF.
FT 295 332  PF07645; EGF_CA.
FT 295 333
PF07645; EGF_CA.
FT 295 333  PS50026; EGF_3.
FT 295 333
PS50026; EGF_3.
FT 295 333  SM00179; EGF_CA.
FT 298 333
SM00179; EGF_CA.
FT 298 333  SM00181; EGF.
FT 335 371
SM00181; EGF.
FT 335 371  PS50026; EGF_3.
FT 335 371
PS50026; EGF_3.
FT 335 371  SM00179; EGF_CA.
FT 338 371
SM00179; EGF_CA.
FT 338 371  SM00181; EGF.
FT 339 370
SM00181; EGF.
FT 339 370  PF00008; EGF.
FT 372 410
PF00008; EGF.
FT 372 410  PS50026; EGF_3.
FT 375 410
PS50026; EGF_3.
FT 375 410  SM00181; EGF.
FT 376 409
SM00181; EGF.
FT 376 409  PF00008; EGF.
FT 412 449
PF00008; EGF.
FT 412 449  PF07645; EGF_CA.
FT 412 450
PF07645; EGF_CA.
FT 412 450  PS50026; EGF_3.
FT 412 450
PS50026; EGF_3.
FT 412 450  SM00179; EGF_CA.
FT 415 450
SM00179; EGF_CA.
FT 415 450  SM00181; EGF.
FT 452 488
SM00181; EGF.
FT 452 488  PS50026; EGF_3.
FT 452 488
PS50026; EGF_3.
FT 452 488  SM00179; EGF_CA.
FT 455 488
SM00179; EGF_CA.
FT 455 488  SM00181; EGF.
FT 456 487
SM00181; EGF.
FT 456 487  PF00008; EGF.
FT 490 526
PF00008; EGF.
FT 490 526  PS50026; EGF_3.
FT 490 526
PS50026; EGF_3.
FT 490 526  SM00179; EGF_CA.
FT 493 526
SM00179; EGF_CA.
FT 493 526  SM00181; EGF.
FT 494 525
SM00181; EGF.
FT 494 525  PF00008; EGF.
FT 528 564
PF00008; EGF.
FT 528 564  PS50026; EGF_3.
FT 528 564
PS50026; EGF_3.
FT 528 564  SM00179; EGF_CA.
FT 531 564
SM00179; EGF_CA.
FT 531 564  SM00181; EGF.
FT 532 563
SM00181; EGF.
FT 532 563  PF00008; EGF.
FT 566 601
PF00008; EGF.
FT 566 601  PS50026; EGF_3.
FT 566 601
PS50026; EGF_3.
FT 566 601  SM00179; EGF_CA.
FT 569 601
SM00179; EGF_CA.
FT 569 601  SM00181; EGF.
FT 570 600
SM00181; EGF.
FT 570 600  PF00008; EGF.
FT 603 639
PF00008; EGF.
FT 603 639  PS50026; EGF_3.
FT 603 639
PS50026; EGF_3.
FT 603 639  SM00179; EGF_CA.
FT 606 639
SM00179; EGF_CA.
FT 606 639  SM00181; EGF.
FT 607 638
SM00181; EGF.
FT 607 638  PF00008; EGF.
FT 641 676
PF00008; EGF.
FT 641 676  PS50026; EGF_3.
FT 641 676
PS50026; EGF_3.
FT 641 676  SM00179; EGF_CA.
FT 644 676
SM00179; EGF_CA.
FT 644 676  SM00181; EGF.
FT 645 675
SM00181; EGF.
FT 645 675  PF00008; EGF.
FT 678 714
PF00008; EGF.
FT 678 714  PS50026; EGF_3.
FT 678 714
PS50026; EGF_3.
FT 678 714  SM00179; EGF_CA.
FT 681 714
SM00179; EGF_CA.
FT 681 714  SM00181; EGF.
FT 682 713
SM00181; EGF.
FT 682 713  PF00008; EGF.
FT 716 751
PF00008; EGF.
FT 716 751  PS50026; EGF_3.
FT 716 751
PS50026; EGF_3.
FT 716 751  SM00179; EGF_CA.
FT 719 751
SM00179; EGF_CA.
FT 719 751  SM00181; EGF.
FT 720 750
SM00181; EGF.
FT 720 750  PF00008; EGF.
FT 753 789
PF00008; EGF.
FT 753 789  PS50026; EGF_3.
FT 753 789
PS50026; EGF_3.
FT 753 789  SM00179; EGF_CA.
FT 756 789
SM00179; EGF_CA.
FT 756 789  SM00181; EGF.
FT 757 788
SM00181; EGF.
FT 757 788  PF00008; EGF.
FT 791 827
PF00008; EGF.
FT 791 827  PS50026; EGF_3.
FT 791 827
PS50026; EGF_3.
FT 791 827  SM00179; EGF_CA.
FT 794 827
SM00179; EGF_CA.
FT 794 827  SM00181; EGF.
FT 795 826
SM00181; EGF.
FT 795 826  PF00008; EGF.
FT 829 867
PF00008; EGF.
FT 829 867  PS50026; EGF_3.
FT 832 867
PS50026; EGF_3.
FT 832 867  SM00181; EGF.
FT 833 866
SM00181; EGF.
FT 833 866  PF00008; EGF.
FT 833 867
PF00008; EGF.
FT 833 867  SM00179; EGF_CA.
FT 869 904
SM00179; EGF_CA.
FT 869 904  PF07645; EGF_CA.
FT 869 905
PF07645; EGF_CA.
FT 869 905  PS50026; EGF_3.
FT 869 905
PS50026; EGF_3.
FT 869 905  SM00179; EGF_CA.
FT 872 905
SM00179; EGF_CA.
FT 872 905  SM00181; EGF.
FT 907 943
SM00181; EGF.
FT 907 943  PS50026; EGF_3.
FT 907 943
PS50026; EGF_3.
FT 907 943  SM00179; EGF_CA.
FT 910 943
SM00179; EGF_CA.
FT 910 943  SM00181; EGF.
FT 911 942
SM00181; EGF.
FT 911 942  PF00008; EGF.
FT 945 981
PF00008; EGF.
FT 945 981  PS50026; EGF_3.
FT 945 981
PS50026; EGF_3.
FT 945 981  SM00179; EGF_CA.
FT 948 981
SM00179; EGF_CA.
FT 948 981  SM00181; EGF.
FT 949 980
SM00181; EGF.
FT 949 980  PF00008; EGF.
FT 983 1019
PF00008; EGF.
FT 983 1019  PS50026; EGF_3.
FT 983 1019
PS50026; EGF_3.
FT 983 1019  SM00179; EGF_CA.
FT 986 1019
SM00179; EGF_CA.
FT 986 1019  SM00181; EGF.
FT 987 1018
SM00181; EGF.
FT 987 1018  PF00008; EGF.
FT 1021 1057
PF00008; EGF.
FT 1021 1057  PS50026; EGF_3.
FT 1021 1057
PS50026; EGF_3.
FT 1021 1057  SM00179; EGF_CA.
FT 1024 1057
SM00179; EGF_CA.
FT 1024 1057  SM00181; EGF.
FT 1025 1056
SM00181; EGF.
FT 1025 1056  PF00008; EGF.
FT 1059 1095
PF00008; EGF.
FT 1059 1095  PS50026; EGF_3.
FT 1062 1095
PS50026; EGF_3.
FT 1062 1095  SM00181; EGF.
FT 1063 1094
SM00181; EGF.
FT 1063 1094  PF00008; EGF.
FT 1063 1095
PF00008; EGF.
FT 1063 1095  SM00179; EGF_CA.
FT 1100 1143
SM00179; EGF_CA.
FT 1100 1143  SM00181; EGF.
FT 1101 1143
SM00181; EGF.
FT 1101 1143  SM00179; EGF_CA.
FT 1108 1143
SM00179; EGF_CA.
FT 1108 1143  PS50026; EGF_3.
FT 1116 1142
PS50026; EGF_3.
FT 1116 1142  PF00008; EGF.
FT 1145 1181
PF00008; EGF.
FT 1145 1181  PS50026; EGF_3.
FT 1145 1181
PS50026; EGF_3.
FT 1145 1181  SM00179; EGF_CA.
FT 1148 1181
SM00179; EGF_CA.
FT 1148 1181  SM00181; EGF.
FT 1149 1180
SM00181; EGF.
FT 1149 1180  PF00008; EGF.
FT 1183 1219
PF00008; EGF.
FT 1183 1219  PS50026; EGF_3.
FT 1183 1219
PS50026; EGF_3.
FT 1183 1219  SM00179; EGF_CA.
FT 1186 1219
SM00179; EGF_CA.
FT 1186 1219  SM00181; EGF.
FT 1187 1218
SM00181; EGF.
FT 1187 1218  PF00008; EGF.
FT 1221 1265
PF00008; EGF.
FT 1221 1265  PS50026; EGF_3.
FT 1221 1265
PS50026; EGF_3.
FT 1221 1265  SM00179; EGF_CA.
FT 1224 1265
SM00179; EGF_CA.
FT 1224 1265  SM00181; EGF.
FT 1238 1264
SM00181; EGF.
FT 1238 1264  PF00008; EGF.
FT 1267 1305
PF00008; EGF.
FT 1267 1305  PS50026; EGF_3.
FT 1267 1305
PS50026; EGF_3.
FT 1267 1305  SM00179; EGF_CA.
FT 1270 1305
SM00179; EGF_CA.
FT 1270 1305  SM00181; EGF.
FT 1271 1304
SM00181; EGF.
FT 1271 1304  PF00008; EGF.
FT 1307 1346
PF00008; EGF.
FT 1307 1346  PS50026; EGF_3.
FT 1308 1346
PS50026; EGF_3.
FT 1308 1346  SM00179; EGF_CA.
FT 1310 1346
SM00179; EGF_CA.
FT 1310 1346  SM00181; EGF.
FT 1311 1345
SM00181; EGF.
FT 1311 1345  PF00008; EGF.
FT 1348 1384
PF00008; EGF.
FT 1348 1384  PS50026; EGF_3.
FT 1348 1384
PS50026; EGF_3.
FT 1348 1384  SM00179; EGF_CA.
FT 1351 1384
SM00179; EGF_CA.
FT 1351 1384  SM00181; EGF.
FT 1352 1383
SM00181; EGF.
FT 1352 1383  PF00008; EGF.
FT 1387 1426
PF00008; EGF.
FT 1387 1426  PS50026; EGF_3.
FT 1390 1426
PS50026; EGF_3.
FT 1390 1426  SM00181; EGF.
FT 1391 1425
SM00181; EGF.
FT 1391 1425  PF00008; EGF.
FT 1442 1480
PF00008; EGF.
FT 1442 1480  PF00066; Notch.
FT 1442 1480
PF00066; Notch.
FT 1442 1480  SM00004; NL.
FT 1449 1489
SM00004; NL.
FT 1449 1489  PS50258; LNR.
FT 1483 1522
PS50258; LNR.
FT 1483 1522  SM00004; NL.
FT 1485 1522
SM00004; NL.
FT 1485 1522  PF00066; Notch.
FT 1490 1531
PF00066; Notch.
FT 1490 1531  PS50258; LNR.
FT 1523 1562
PS50258; LNR.
FT 1523 1562  SM00004; NL.
FT 1524 1562
SM00004; NL.
FT 1524 1562  PF00066; Notch.
FT 1532 1571
PF00066; Notch.
FT 1532 1571  PS50258; LNR.
FT 1566 1622
PS50258; LNR.
FT 1566 1622  PF06816; NOD.
FT 1660 1722
PF06816; NOD.
FT 1660 1722  PF07684; NODP.
FT 1863 2102
PF07684; NODP.
FT 1863 2102  PS50297; ANK_REP_REGION.
FT 1870 1912
PS50297; ANK_REP_REGION.
FT 1870 1912  SM00248; ANK.
FT 1870 1915
SM00248; ANK.
FT 1870 1915  PF00023; Ank.
FT 1917 1932
PF00023; Ank.
FT 1917 1932  PF00023; Ank.
FT 1917 1947
PF00023; Ank.
FT 1917 1947  SM00248; ANK.
FT 1937 1948
SM00248; ANK.
FT 1937 1948  PF00023; Ank.
FT 1949 1979
PF00023; Ank.
FT 1949 1979  SM00248; ANK.
FT 1949 1982
SM00248; ANK.
FT 1949 1982  PF00023; Ank.
FT 1983 2012
PF00023; Ank.
FT 1983 2012  SM00248; ANK.
FT 1983 2015
SM00248; ANK.
FT 1983 2015  PF00023; Ank.
FT 1983 2015
PF00023; Ank.
FT 1983 2015  PS50088; ANK_REPEAT.
FT 2016 2045
PS50088; ANK_REPEAT.
FT 2016 2045  SM00248; ANK.
FT 2016 2048
SM00248; ANK.
FT 2016 2048  PF00023; Ank.
FT 2016 2048
PF00023; Ank.
FT 2016 2048  PS50088; ANK_REPEAT.
FT 2049 2078
PS50088; ANK_REPEAT.
FT 2049 2078  SM00248; ANK.
FT 2049 2081
SM00248; ANK.
FT 2049 2081  PF00023; Ank.
FT 2194 2398
PF00023; Ank.
FT 2194 2398  TAD (transactivation domain) [2].
TAD (transactivation domain) [2].
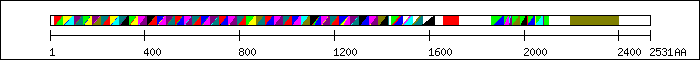 XX
IN T09547 MEF-2C; mouse, Mus musculus.
XX
DR TRANSPATH: MO000085514.
DR EMBL: Z11886;
DR UniProtKB: Q01705-1;
XX
RN [1]; RE0015210.
RX PUBMED: 10082551.
RA Wilson-Rawls J., Molkentin J. D., Black B. L., Olson E. N.
RT Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C
RL Mol. Cell. Biol. 19:2853-2862 (1999).
RN [2]; RE0021256.
RX PUBMED: 9826771.
RA Kurooka H., Kuroda K., Honjo T.
RT Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region
RL Nucleic Acids Res. 26:5448-5455 (1998).
XX
//
XX
IN T09547 MEF-2C; mouse, Mus musculus.
XX
DR TRANSPATH: MO000085514.
DR EMBL: Z11886;
DR UniProtKB: Q01705-1;
XX
RN [1]; RE0015210.
RX PUBMED: 10082551.
RA Wilson-Rawls J., Molkentin J. D., Black B. L., Olson E. N.
RT Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C
RL Mol. Cell. Biol. 19:2853-2862 (1999).
RN [2]; RE0021256.
RX PUBMED: 9826771.
RA Kurooka H., Kuroda K., Honjo T.
RT Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region
RL Nucleic Acids Res. 26:5448-5455 (1998).
XX
//


PS50026; EGF_3. FT 23 58
SM00181; EGF. FT 59 99
PS50026; EGF_3. FT 59 99
SM00179; EGF_CA. FT 62 99
SM00181; EGF. FT 63 98
PF00008; EGF. FT 102 139
PS50026; EGF_3. FT 105 139
SM00181; EGF. FT 106 138
PF00008; EGF. FT 106 139
SM00179; EGF_CA. FT 140 176
PS50026; EGF_3. FT 140 176
SM00179; EGF_CA. FT 143 176
SM00181; EGF. FT 144 175
PF00008; EGF. FT 178 215
PF07645; EGF_CA. FT 178 216
PS50026; EGF_3. FT 178 216
SM00179; EGF_CA. FT 181 216
SM00181; EGF. FT 218 255
PS50026; EGF_3. FT 221 255
SM00181; EGF. FT 222 254
PF00008; EGF. FT 222 255
SM00179; EGF_CA. FT 257 293
PS50026; EGF_3. FT 257 293
SM00179; EGF_CA. FT 260 293
SM00181; EGF. FT 261 292
PF00008; EGF. FT 295 332
PF07645; EGF_CA. FT 295 333
PS50026; EGF_3. FT 295 333
SM00179; EGF_CA. FT 298 333
SM00181; EGF. FT 335 371
PS50026; EGF_3. FT 335 371
SM00179; EGF_CA. FT 338 371
SM00181; EGF. FT 339 370
PF00008; EGF. FT 372 410
PS50026; EGF_3. FT 375 410
SM00181; EGF. FT 376 409
PF00008; EGF. FT 412 449
PF07645; EGF_CA. FT 412 450
PS50026; EGF_3. FT 412 450
SM00179; EGF_CA. FT 415 450
SM00181; EGF. FT 452 488
PS50026; EGF_3. FT 452 488
SM00179; EGF_CA. FT 455 488
SM00181; EGF. FT 456 487
PF00008; EGF. FT 490 526
PS50026; EGF_3. FT 490 526
SM00179; EGF_CA. FT 493 526
SM00181; EGF. FT 494 525
PF00008; EGF. FT 528 564
PS50026; EGF_3. FT 528 564
SM00179; EGF_CA. FT 531 564
SM00181; EGF. FT 532 563
PF00008; EGF. FT 566 601
PS50026; EGF_3. FT 566 601
SM00179; EGF_CA. FT 569 601
SM00181; EGF. FT 570 600
PF00008; EGF. FT 603 639
PS50026; EGF_3. FT 603 639
SM00179; EGF_CA. FT 606 639
SM00181; EGF. FT 607 638
PF00008; EGF. FT 641 676
PS50026; EGF_3. FT 641 676
SM00179; EGF_CA. FT 644 676
SM00181; EGF. FT 645 675
PF00008; EGF. FT 678 714
PS50026; EGF_3. FT 678 714
SM00179; EGF_CA. FT 681 714
SM00181; EGF. FT 682 713
PF00008; EGF. FT 716 751
PS50026; EGF_3. FT 716 751
SM00179; EGF_CA. FT 719 751
SM00181; EGF. FT 720 750
PF00008; EGF. FT 753 789
PS50026; EGF_3. FT 753 789
SM00179; EGF_CA. FT 756 789
SM00181; EGF. FT 757 788
PF00008; EGF. FT 791 827
PS50026; EGF_3. FT 791 827
SM00179; EGF_CA. FT 794 827
SM00181; EGF. FT 795 826
PF00008; EGF. FT 829 867
PS50026; EGF_3. FT 832 867
SM00181; EGF. FT 833 866
PF00008; EGF. FT 833 867
SM00179; EGF_CA. FT 869 904
PF07645; EGF_CA. FT 869 905
PS50026; EGF_3. FT 869 905
SM00179; EGF_CA. FT 872 905
SM00181; EGF. FT 907 943
PS50026; EGF_3. FT 907 943
SM00179; EGF_CA. FT 910 943
SM00181; EGF. FT 911 942
PF00008; EGF. FT 945 981
PS50026; EGF_3. FT 945 981
SM00179; EGF_CA. FT 948 981
SM00181; EGF. FT 949 980
PF00008; EGF. FT 983 1019
PS50026; EGF_3. FT 983 1019
SM00179; EGF_CA. FT 986 1019
SM00181; EGF. FT 987 1018
PF00008; EGF. FT 1021 1057
PS50026; EGF_3. FT 1021 1057
SM00179; EGF_CA. FT 1024 1057
SM00181; EGF. FT 1025 1056
PF00008; EGF. FT 1059 1095
PS50026; EGF_3. FT 1062 1095
SM00181; EGF. FT 1063 1094
PF00008; EGF. FT 1063 1095
SM00179; EGF_CA. FT 1100 1143
SM00181; EGF. FT 1101 1143
SM00179; EGF_CA. FT 1108 1143
PS50026; EGF_3. FT 1116 1142
PF00008; EGF. FT 1145 1181
PS50026; EGF_3. FT 1145 1181
SM00179; EGF_CA. FT 1148 1181
SM00181; EGF. FT 1149 1180
PF00008; EGF. FT 1183 1219
PS50026; EGF_3. FT 1183 1219
SM00179; EGF_CA. FT 1186 1219
SM00181; EGF. FT 1187 1218
PF00008; EGF. FT 1221 1265
PS50026; EGF_3. FT 1221 1265
SM00179; EGF_CA. FT 1224 1265
SM00181; EGF. FT 1238 1264
PF00008; EGF. FT 1267 1305
PS50026; EGF_3. FT 1267 1305
SM00179; EGF_CA. FT 1270 1305
SM00181; EGF. FT 1271 1304
PF00008; EGF. FT 1307 1346
PS50026; EGF_3. FT 1308 1346
SM00179; EGF_CA. FT 1310 1346
SM00181; EGF. FT 1311 1345
PF00008; EGF. FT 1348 1384
PS50026; EGF_3. FT 1348 1384
SM00179; EGF_CA. FT 1351 1384
SM00181; EGF. FT 1352 1383
PF00008; EGF. FT 1387 1426
PS50026; EGF_3. FT 1390 1426
SM00181; EGF. FT 1391 1425
PF00008; EGF. FT 1442 1480
PF00066; Notch. FT 1442 1480
SM00004; NL. FT 1449 1489
PS50258; LNR. FT 1483 1522
SM00004; NL. FT 1485 1522
PF00066; Notch. FT 1490 1531
PS50258; LNR. FT 1523 1562
SM00004; NL. FT 1524 1562
PF00066; Notch. FT 1532 1571
PS50258; LNR. FT 1566 1622
PF06816; NOD. FT 1660 1722
PF07684; NODP. FT 1863 2102
PS50297; ANK_REP_REGION. FT 1870 1912
SM00248; ANK. FT 1870 1915
PF00023; Ank. FT 1917 1932
PF00023; Ank. FT 1917 1947
SM00248; ANK. FT 1937 1948
PF00023; Ank. FT 1949 1979
SM00248; ANK. FT 1949 1982
PF00023; Ank. FT 1983 2012
SM00248; ANK. FT 1983 2015
PF00023; Ank. FT 1983 2015
PS50088; ANK_REPEAT. FT 2016 2045
SM00248; ANK. FT 2016 2048
PF00023; Ank. FT 2016 2048
PS50088; ANK_REPEAT. FT 2049 2078
SM00248; ANK. FT 2049 2081
PF00023; Ank. FT 2194 2398
TAD (transactivation domain) [2].
XX IN T09547 MEF-2C; mouse, Mus musculus. XX DR TRANSPATH: MO000085514. DR EMBL: Z11886; DR UniProtKB: Q01705-1; XX RN [1]; RE0015210. RX PUBMED: 10082551. RA Wilson-Rawls J., Molkentin J. D., Black B. L., Olson E. N. RT Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C RL Mol. Cell. Biol. 19:2853-2862 (1999). RN [2]; RE0021256. RX PUBMED: 9826771. RA Kurooka H., Kuroda K., Honjo T. RT Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region RL Nucleic Acids Res. 26:5448-5455 (1998). XX //