
AC T02144
XX
ID T02144
XX
DT 26.05.1997 (created); hiwi.
DT 29.11.2012 (updated); mkl.
CO Copyright (C), QIAGEN.
XX
FA Ada2p
XX
SY ADA2; ADA2p.
XX
OS yeast, Saccharomyces cerevisiae
OC Eukaryota; Fungi; Ascomycota; Hemiascomycetes; Saccharomycetales; Saccharomycetaceae; Saccharomyces.
XX
GE G004229 ADA2.
XX
HO Ada2p (human).
XX
SZ 434 AA; 50.6 kDa (cDNA) (calc.).
XX
SQ MSNKFHCDVCSADCTNRVRVSCAICPEYDLCVPCFSQGSYTGKHRPYHDYRIIETNSYPI
SQ LCPDWGADEELQLIKGAQTLGLGNWQDIADHIGSRGKEEVKEHYLKYYLESKYYPIPDIT
SQ QNIHVPQDEFLEQRRHRIESFRERPLEPPRKPMASVPSCHEVQGFMPGRLEFETEFENEA
SQ EGPVKDMVFEPDDQPLDIELKFAILDIYNSRLTTRAEKKRLLFENHLMDYRKLQAIDKKR
SQ SKEAKELYNRIKPFARVMTAQDFEEFSKDILEELHCRARIQQLQEWRSNGLTTLEAGLKY
SQ ERDKQARISSFEKFGASTAASLSEGNSRYRSNSAHRSNAEYSQNYSENGGRKKNMTISDI
SQ QHAPDYALLSNDEQQLCIQLKILPKPYLVLKEVMFRELLKTGGNLSKSACRELLNIDPIK
SQ ANRIYDFFQSQNWM
XX
SC Swiss-Prot#Q02336
XX
FT 1 46  PF00569; Zinc finger, ZZ type.
FT 1 46
PF00569; Zinc finger, ZZ type.
FT 1 46  SM00291; zz_5.
FT 1 48
SM00291; zz_5.
FT 1 48  PS50135; ZF_ZZ_2.
FT 61 110
PS50135; ZF_ZZ_2.
FT 61 110  SM00717; sant.
FT 62 108
SM00717; sant.
FT 62 108  PF00249; Myb-like DNA-binding domain.
FT 65 95
PF00249; Myb-like DNA-binding domain.
FT 65 95  c-Myb (30-60) homology region [2].
FT 65 108
c-Myb (30-60) homology region [2].
FT 65 108  PS50090; MYB_3.
FT 349 434
PS50090; MYB_3.
FT 349 434  PF04433; SWIRM domain.
FT 349 434
PF04433; SWIRM domain.
FT 349 434  PS50934; SWIRM.
PS50934; SWIRM.
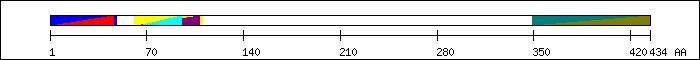 XX
SF similarity with a single Myb-like tryptophan repeat [2];
SF N-terminal part interacts directly with the acidic trans-activation domain of Vmw65 T00894 [1];
XX
FF coactivator / adaptor (as complex with GCN5 T02145) for certain acidic activation domains [2] [5];
FF may have to cooperate with ADA3 [4];
XX
IN T02145 Gcn5p; yeast, Saccharomyces cerevisiae.
IN T00894 Vmw65; HSV-1, herpes simplex virus type 1.
XX
DR EMBL: M95396;
DR EMBL: U33007;
DR UniProtKB: Q02336;
DR UniProtKB: Q02354;
XX
RN [1]; RE0005636.
RX PUBMED: 7642611.
RA Barlev N. A., Candau R., Wang L., Darpino P., Silverman N., Berger S. L.
RT Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein
RL J. Biol. Chem. 270:19337-19344 (1995).
RN [2]; RE0005637.
RX PUBMED: 1638630.
RA Berger S. L., Pina B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L.
RT Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains
RL Cell 70:251-265 (1992).
RN [3]; RE0005638.
RX PUBMED: 7957049.
RA Marcus G. A., Silverman N., Berger S. L., Horiuchi J., Guarente L.
RT Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors
RL EMBO J. 13:4807-4815 (1994).
RN [4]; RE0005639.
RX PUBMED: 7972120.
RA Silverman N., Agapite J., Guarente L.
RT Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription
RL Proc. Natl. Acad. Sci. USA 91:11665-11668 (1994).
RN [5]; RE0005640.
RX PUBMED: 1396595.
RA Georgakopoulos T., Thireos G.
RT Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription
RL EMBO J. 11:4145-4152 (1992).
XX
//
XX
SF similarity with a single Myb-like tryptophan repeat [2];
SF N-terminal part interacts directly with the acidic trans-activation domain of Vmw65 T00894 [1];
XX
FF coactivator / adaptor (as complex with GCN5 T02145) for certain acidic activation domains [2] [5];
FF may have to cooperate with ADA3 [4];
XX
IN T02145 Gcn5p; yeast, Saccharomyces cerevisiae.
IN T00894 Vmw65; HSV-1, herpes simplex virus type 1.
XX
DR EMBL: M95396;
DR EMBL: U33007;
DR UniProtKB: Q02336;
DR UniProtKB: Q02354;
XX
RN [1]; RE0005636.
RX PUBMED: 7642611.
RA Barlev N. A., Candau R., Wang L., Darpino P., Silverman N., Berger S. L.
RT Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein
RL J. Biol. Chem. 270:19337-19344 (1995).
RN [2]; RE0005637.
RX PUBMED: 1638630.
RA Berger S. L., Pina B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L.
RT Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains
RL Cell 70:251-265 (1992).
RN [3]; RE0005638.
RX PUBMED: 7957049.
RA Marcus G. A., Silverman N., Berger S. L., Horiuchi J., Guarente L.
RT Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors
RL EMBO J. 13:4807-4815 (1994).
RN [4]; RE0005639.
RX PUBMED: 7972120.
RA Silverman N., Agapite J., Guarente L.
RT Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription
RL Proc. Natl. Acad. Sci. USA 91:11665-11668 (1994).
RN [5]; RE0005640.
RX PUBMED: 1396595.
RA Georgakopoulos T., Thireos G.
RT Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription
RL EMBO J. 11:4145-4152 (1992).
XX
//


PF00569; Zinc finger, ZZ type. FT 1 46
SM00291; zz_5. FT 1 48
PS50135; ZF_ZZ_2. FT 61 110
SM00717; sant. FT 62 108
PF00249; Myb-like DNA-binding domain. FT 65 95
c-Myb (30-60) homology region [2]. FT 65 108
PS50090; MYB_3. FT 349 434
PF04433; SWIRM domain. FT 349 434
PS50934; SWIRM.
XX SF similarity with a single Myb-like tryptophan repeat [2]; SF N-terminal part interacts directly with the acidic trans-activation domain of Vmw65 T00894 [1]; XX FF coactivator / adaptor (as complex with GCN5 T02145) for certain acidic activation domains [2] [5]; FF may have to cooperate with ADA3 [4]; XX IN T02145 Gcn5p; yeast, Saccharomyces cerevisiae. IN T00894 Vmw65; HSV-1, herpes simplex virus type 1. XX DR EMBL: M95396; DR EMBL: U33007; DR UniProtKB: Q02336; DR UniProtKB: Q02354; XX RN [1]; RE0005636. RX PUBMED: 7642611. RA Barlev N. A., Candau R., Wang L., Darpino P., Silverman N., Berger S. L. RT Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein RL J. Biol. Chem. 270:19337-19344 (1995). RN [2]; RE0005637. RX PUBMED: 1638630. RA Berger S. L., Pina B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L. RT Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains RL Cell 70:251-265 (1992). RN [3]; RE0005638. RX PUBMED: 7957049. RA Marcus G. A., Silverman N., Berger S. L., Horiuchi J., Guarente L. RT Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors RL EMBO J. 13:4807-4815 (1994). RN [4]; RE0005639. RX PUBMED: 7972120. RA Silverman N., Agapite J., Guarente L. RT Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription RL Proc. Natl. Acad. Sci. USA 91:11665-11668 (1994). RN [5]; RE0005640. RX PUBMED: 1396595. RA Georgakopoulos T., Thireos G. RT Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription RL EMBO J. 11:4145-4152 (1992). XX //