
AC T00523
XX
ID T00523
XX
DT 21.10.1992 (created); ewi.
DT 02.06.2010 (updated); ada.
CO Copyright (C), QIAGEN.
XX
FA Max
XX
SY Myc-binding novel HLH/LZ protein; Myn.
XX
OS mouse, Mus musculus
OC eukaryota; animalia; metazoa; chordata; vertebrata; tetrapoda; mammalia; eutheria; rodentia; myomorpha; muridae; murinae
XX
GE G006414 Max.
XX
CL C0012; bHLH-ZIP; 1.2.6.5.5.1.
XX
SZ 160 AA; 18.2 kDa (cDNA) (calc.), 20 kDa, 22 kDa (SDS)
XX
SQ MSDNDDIEVESDEEQARFQSAADKRAHHNALERKRRDHIKDSFHSLRDSVPSLQGEKASR
SQ AQILDKATEYIQYMRRKNDTHQQDIDDLKRQNALLEQQVRALEKARSSAQLQTNYPSSDN
SQ SLYTNAKGGTISAFDGGSDSSSESEPEEPQSRKKLRMEAS
XX
SC Swiss-Prot#P28574
XX
FT 2 2  Ser-phosphorylation by CKII [2].
FT 11 11
Ser-phosphorylation by CKII [2].
FT 11 11  Ser-phosphorylation by CKII [2].
FT 24 75
Ser-phosphorylation by CKII [2].
FT 24 75  PF00010; Helix-loop-helix DNA-binding domain.
FT 24 75
PF00010; Helix-loop-helix DNA-binding domain.
FT 24 75  PS50888; HLH.
FT 29 80
PS50888; HLH.
FT 29 80  SM00353; finulus.
SM00353; finulus.
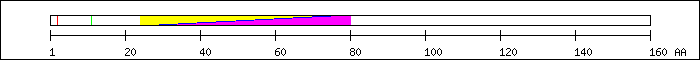 XX
SF solution of the crystal structure revealed that the HLH motifs form a four-helix-bundle with all helices in parallel orientation [5];
SF helix 2 is prolonged by the alpha-helically folded leucine zipper;
SF heterodimers with c-Myc or Mad have higher DNA-binding affinity [1];
SF the leucine zipper is specifically required for heterodimerization [1];
SF belongs to group B HLH-proteins that bind to CACGTG motif in DNA which is due to certain residues in DNA-binding domain (His at pos. 28, Glu at pos. 32, Arg at pos. 36) [8] [9];
XX
CP newborn: intestine, weak in brain, liver, lung; adult: very strong in intestine, weak in brain, kidney, lung [7].
CN newborn: kidney; adult: liver [7].
XX
FF enhances c-Myc DNA-binding, perturbation of homodimer-binding after CKII phosphorylation, but not of Myc/Max-heterodimer [2];
FF in complexes with either Mxi1 or Mad, it represses c-Myc or N-Myc effects [4];
FF murine homolog of MAX [1];
XX
IN T00140 c-Myc-isoform1; human, Homo sapiens.
IN T00141 c-Myc; chick, Gallus gallus.
IN T00142 c-Myc; rat, Rattus norvegicus.
IN T00143 c-Myc; mouse, Mus musculus.
IN T00523 Max; mouse, Mus musculus.
IN T01445 N-Myc; mouse, Mus musculus.
IN T02379 N-Myc; human, Homo sapiens.
XX
MX M01034 V$EBOX_Q6_01.
MX M00119 V$MAX_01.
MX M02881 V$MAX_04.
MX M01830 V$MAX_Q6.
MX M00118 V$MYCMAX_01.
MX M00123 V$MYCMAX_02.
MX M00615 V$MYCMAX_03.
MX M00322 V$MYCMAX_B.
MX M00799 V$MYC_Q2.
XX
BS R02094.
BS R04455.
XX
DR TRANSPATH: MO000024987.
DR EMBL: M63903;
DR UniProtKB: P28574;
XX
RN [1]; RE0000173.
RX PUBMED: 1840505.
RA Prendergast G. C., Lawe D., Ziff E. B.
RT Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation
RL Cell 65:395-407 (1991).
RN [2]; RE0000705.
RX PUBMED: 1737614.
RA Berberich S. J., Cole M. D.
RT Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers
RL Genes Dev. 6:166-176 (1992).
RN [3]; RE0003351.
RX PUBMED: 1644290.
RA Mukherjee B., Morgenbesser S. D., DePinho R. A.
RT Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants
RL Genes Dev. 6:1480-1492 (1992).
RN [4]; RE0003363.
RX PUBMED: 8202517.
RA Lahoz E. G., Xu L., Schreiber-Agus N., DePinho R. A.
RT Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1
RL Proc. Natl. Acad. Sci. USA 91:5503-5507 (1994).
RN [5]; RE0003393.
RX PUBMED: 8479534.
RA Ferre-D'Amare A. E., Prendergast G. C., Ziff E. B., Burley S. K.
RT Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain
RL Nature 363:38-45 (1993).
RN [6]; RE0003395.
RX PUBMED: 1459463.
RA Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B.
RT Biphasic effect of Max and Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions
RL Genes Dev. 6:2429-2439 (1992).
RN [7]; RE0005851.
RX PUBMED: 7667316.
RA Chin L., Schreiber-Agus N., Pellicer I., Chen K., Lee H.-W., Dudast M., Cordon-Cardo C., DePinho R. A.
RT Contrasting roles for Myc and Mad proteins in cellular growth and differentiation
RL Proc. Natl. Acad. Sci. USA 92:8488-8492 (1995).
RN [8]; RE0017805.
RX PUBMED: 9860302.
RA Meng X., Lu X., Li Z., Green E. D., Massa H., Trask B. J., Morris C. A., Keating M. T.
RT Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes
RL Hum. Genet. 103:590-599 (1998).
RN [9]; RE0017817.
RX PUBMED: 11073985.
RA Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E.
RT MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network
RL Mol. Cell. Biol. 20:8845-8854 (2000).
RN [10]; RE0066993.
RX PUBMED: 19843539.
RA Reichman S., Kalathur R. K., Lambard S., Ait-Ali N., Yang Y., Lardenois A., Ripp R., Poch O., Zack D. J., Sahel J. A., Leveillard T.
RT The homeobox gene CHX10/VSX2 regulates RdCVF promoter activity in the inner retina.
RL Hum. Mol. Genet. 19:250-261 (2010).
XX
//
XX
SF solution of the crystal structure revealed that the HLH motifs form a four-helix-bundle with all helices in parallel orientation [5];
SF helix 2 is prolonged by the alpha-helically folded leucine zipper;
SF heterodimers with c-Myc or Mad have higher DNA-binding affinity [1];
SF the leucine zipper is specifically required for heterodimerization [1];
SF belongs to group B HLH-proteins that bind to CACGTG motif in DNA which is due to certain residues in DNA-binding domain (His at pos. 28, Glu at pos. 32, Arg at pos. 36) [8] [9];
XX
CP newborn: intestine, weak in brain, liver, lung; adult: very strong in intestine, weak in brain, kidney, lung [7].
CN newborn: kidney; adult: liver [7].
XX
FF enhances c-Myc DNA-binding, perturbation of homodimer-binding after CKII phosphorylation, but not of Myc/Max-heterodimer [2];
FF in complexes with either Mxi1 or Mad, it represses c-Myc or N-Myc effects [4];
FF murine homolog of MAX [1];
XX
IN T00140 c-Myc-isoform1; human, Homo sapiens.
IN T00141 c-Myc; chick, Gallus gallus.
IN T00142 c-Myc; rat, Rattus norvegicus.
IN T00143 c-Myc; mouse, Mus musculus.
IN T00523 Max; mouse, Mus musculus.
IN T01445 N-Myc; mouse, Mus musculus.
IN T02379 N-Myc; human, Homo sapiens.
XX
MX M01034 V$EBOX_Q6_01.
MX M00119 V$MAX_01.
MX M02881 V$MAX_04.
MX M01830 V$MAX_Q6.
MX M00118 V$MYCMAX_01.
MX M00123 V$MYCMAX_02.
MX M00615 V$MYCMAX_03.
MX M00322 V$MYCMAX_B.
MX M00799 V$MYC_Q2.
XX
BS R02094.
BS R04455.
XX
DR TRANSPATH: MO000024987.
DR EMBL: M63903;
DR UniProtKB: P28574;
XX
RN [1]; RE0000173.
RX PUBMED: 1840505.
RA Prendergast G. C., Lawe D., Ziff E. B.
RT Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation
RL Cell 65:395-407 (1991).
RN [2]; RE0000705.
RX PUBMED: 1737614.
RA Berberich S. J., Cole M. D.
RT Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers
RL Genes Dev. 6:166-176 (1992).
RN [3]; RE0003351.
RX PUBMED: 1644290.
RA Mukherjee B., Morgenbesser S. D., DePinho R. A.
RT Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants
RL Genes Dev. 6:1480-1492 (1992).
RN [4]; RE0003363.
RX PUBMED: 8202517.
RA Lahoz E. G., Xu L., Schreiber-Agus N., DePinho R. A.
RT Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1
RL Proc. Natl. Acad. Sci. USA 91:5503-5507 (1994).
RN [5]; RE0003393.
RX PUBMED: 8479534.
RA Ferre-D'Amare A. E., Prendergast G. C., Ziff E. B., Burley S. K.
RT Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain
RL Nature 363:38-45 (1993).
RN [6]; RE0003395.
RX PUBMED: 1459463.
RA Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B.
RT Biphasic effect of Max and Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions
RL Genes Dev. 6:2429-2439 (1992).
RN [7]; RE0005851.
RX PUBMED: 7667316.
RA Chin L., Schreiber-Agus N., Pellicer I., Chen K., Lee H.-W., Dudast M., Cordon-Cardo C., DePinho R. A.
RT Contrasting roles for Myc and Mad proteins in cellular growth and differentiation
RL Proc. Natl. Acad. Sci. USA 92:8488-8492 (1995).
RN [8]; RE0017805.
RX PUBMED: 9860302.
RA Meng X., Lu X., Li Z., Green E. D., Massa H., Trask B. J., Morris C. A., Keating M. T.
RT Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes
RL Hum. Genet. 103:590-599 (1998).
RN [9]; RE0017817.
RX PUBMED: 11073985.
RA Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E.
RT MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network
RL Mol. Cell. Biol. 20:8845-8854 (2000).
RN [10]; RE0066993.
RX PUBMED: 19843539.
RA Reichman S., Kalathur R. K., Lambard S., Ait-Ali N., Yang Y., Lardenois A., Ripp R., Poch O., Zack D. J., Sahel J. A., Leveillard T.
RT The homeobox gene CHX10/VSX2 regulates RdCVF promoter activity in the inner retina.
RL Hum. Mol. Genet. 19:250-261 (2010).
XX
//


Ser-phosphorylation by CKII [2]. FT 11 11
Ser-phosphorylation by CKII [2]. FT 24 75
PF00010; Helix-loop-helix DNA-binding domain. FT 24 75
PS50888; HLH. FT 29 80
SM00353; finulus.
XX SF solution of the crystal structure revealed that the HLH motifs form a four-helix-bundle with all helices in parallel orientation [5]; SF helix 2 is prolonged by the alpha-helically folded leucine zipper; SF heterodimers with c-Myc or Mad have higher DNA-binding affinity [1]; SF the leucine zipper is specifically required for heterodimerization [1]; SF belongs to group B HLH-proteins that bind to CACGTG motif in DNA which is due to certain residues in DNA-binding domain (His at pos. 28, Glu at pos. 32, Arg at pos. 36) [8] [9]; XX CP newborn: intestine, weak in brain, liver, lung; adult: very strong in intestine, weak in brain, kidney, lung [7]. CN newborn: kidney; adult: liver [7]. XX FF enhances c-Myc DNA-binding, perturbation of homodimer-binding after CKII phosphorylation, but not of Myc/Max-heterodimer [2]; FF in complexes with either Mxi1 or Mad, it represses c-Myc or N-Myc effects [4]; FF murine homolog of MAX [1]; XX IN T00140 c-Myc-isoform1; human, Homo sapiens. IN T00141 c-Myc; chick, Gallus gallus. IN T00142 c-Myc; rat, Rattus norvegicus. IN T00143 c-Myc; mouse, Mus musculus. IN T00523 Max; mouse, Mus musculus. IN T01445 N-Myc; mouse, Mus musculus. IN T02379 N-Myc; human, Homo sapiens. XX MX M01034 V$EBOX_Q6_01. MX M00119 V$MAX_01. MX M02881 V$MAX_04. MX M01830 V$MAX_Q6. MX M00118 V$MYCMAX_01. MX M00123 V$MYCMAX_02. MX M00615 V$MYCMAX_03. MX M00322 V$MYCMAX_B. MX M00799 V$MYC_Q2. XX BS R02094. BS R04455. XX DR TRANSPATH: MO000024987. DR EMBL: M63903; DR UniProtKB: P28574; XX RN [1]; RE0000173. RX PUBMED: 1840505. RA Prendergast G. C., Lawe D., Ziff E. B. RT Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation RL Cell 65:395-407 (1991). RN [2]; RE0000705. RX PUBMED: 1737614. RA Berberich S. J., Cole M. D. RT Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers RL Genes Dev. 6:166-176 (1992). RN [3]; RE0003351. RX PUBMED: 1644290. RA Mukherjee B., Morgenbesser S. D., DePinho R. A. RT Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants RL Genes Dev. 6:1480-1492 (1992). RN [4]; RE0003363. RX PUBMED: 8202517. RA Lahoz E. G., Xu L., Schreiber-Agus N., DePinho R. A. RT Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1 RL Proc. Natl. Acad. Sci. USA 91:5503-5507 (1994). RN [5]; RE0003393. RX PUBMED: 8479534. RA Ferre-D'Amare A. E., Prendergast G. C., Ziff E. B., Burley S. K. RT Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain RL Nature 363:38-45 (1993). RN [6]; RE0003395. RX PUBMED: 1459463. RA Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B. RT Biphasic effect of Max and Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions RL Genes Dev. 6:2429-2439 (1992). RN [7]; RE0005851. RX PUBMED: 7667316. RA Chin L., Schreiber-Agus N., Pellicer I., Chen K., Lee H.-W., Dudast M., Cordon-Cardo C., DePinho R. A. RT Contrasting roles for Myc and Mad proteins in cellular growth and differentiation RL Proc. Natl. Acad. Sci. USA 92:8488-8492 (1995). RN [8]; RE0017805. RX PUBMED: 9860302. RA Meng X., Lu X., Li Z., Green E. D., Massa H., Trask B. J., Morris C. A., Keating M. T. RT Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes RL Hum. Genet. 103:590-599 (1998). RN [9]; RE0017817. RX PUBMED: 11073985. RA Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E. RT MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network RL Mol. Cell. Biol. 20:8845-8854 (2000). RN [10]; RE0066993. RX PUBMED: 19843539. RA Reichman S., Kalathur R. K., Lambard S., Ait-Ali N., Yang Y., Lardenois A., Ripp R., Poch O., Zack D. J., Sahel J. A., Leveillard T. RT The homeobox gene CHX10/VSX2 regulates RdCVF promoter activity in the inner retina. RL Hum. Mol. Genet. 19:250-261 (2010). XX //