
AC T02245
XX
ID T02245
XX
DT 28.10.1997 (created); ili.
DT 11.02.2014 (updated); hna.
CO Copyright (C), QIAGEN.
XX
FA AML1b
XX
SY Acute Myeloid Leukemia 1B; AML1; CBF-alpha 2; CBFA2; PEBP2alphaB; PEBP2alphaB1; RUNX1.
XX
OS human, Homo sapiens
OC eukaryota; animalia; metazoa; chordata; vertebrata; tetrapoda; mammalia; eutheria; primates
XX
GE G003993 RUNX1; HGNC: RUNX1.
XX
CL C0029; runt; 6.4.1.0.2.2.
XX
SZ 453 AA; 48.7 kDa (cDNA) (calc.).
XX
SQ MRIPVDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRSMVEVLADHPG
SQ ELVRTDSPNFLCSVLPTHWRCNKTLPIAFKVVALGDVPDGTLVTVMAGNDENYSAELRNA
SQ TAAMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPPQVATYHRAIKITVDGPREPRRHR
SQ QKLDDQTKPGSLSFSERLSELEQLRRTAMRVSPHHPAPTPNPRASLNHSTAFNPQPQSQM
SQ QDTRQIQPSPPWSYDQSYQYLGSIASPSVHPATPISPGRASGMTTLSAELSSRLSTAPDL
SQ TAFSDPRQFPALPSISDPRMHYPGAFTYSPTPVTSGIGIGMSAMGSATRYHTYLPPPYPG
SQ SSQAQGGPFQASSPSYHLYYGASAGSYQFSMVGGERSPPRILPPCTNASTGSALLNPSLP
SQ NQSDVVEAEGSHSNSPTNMAPSARLEEAVWRPY
XX
SC Swiss-Prot#Q01196-1
XX
FT 48 182  PF00853; Runt domain.
FT 50 178
PF00853; Runt domain.
FT 50 178  PS51062; RUNT.
FT 66 438
PS51062; RUNT.
FT 66 438  PF00478; IMP dehydrogenase / GMP reductase domain.
FT 72 72
PF00478; IMP dehydrogenase / GMP reductase domain.
FT 72 72  C is important for DNA-binding and biological activity [5].
FT 249 249
C is important for DNA-binding and biological activity [5].
FT 249 249  major extracellular signal-regulated kinase (ERK)-dependent phosphorylation site [4].
FT 266 266
major extracellular signal-regulated kinase (ERK)-dependent phosphorylation site [4].
FT 266 266  minor ERK-dependent phosphorylation site [4].
FT 324 354
minor ERK-dependent phosphorylation site [4].
FT 324 354  putative nuclear matrix targeting signal [6].
FT 392 392
putative nuclear matrix targeting signal [6].
FT 392 392  S replaced by F [3].
S replaced by F [3].
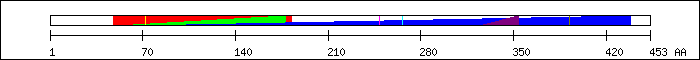 XX
SF very similar to PEBP2alphaB from mouse [1];
SF product of AML1 gene as the result of differential usage of polyadenylation signals [1];
SF related products are AML1a T02256 and AML1c T02246 [1];
SF harbors free SH-groups [5];
XX
CN brain [1], heart [1] [1].
XX
FF nuclear extracts from HTR-8/SV*NEO, JEG-3 and embryonic day 14 placentas were all positive for Runx1b (AML1b), the active form of RUNX [10];
FF expression level of AML1b vary between different tissues [1];
FF related to t(8,21) acute myeloid leukemia;
FF binds to PEBP2 site with lower affinity than AML1a [2];
FF AML1b-trans-activation is negatively regulated by AML1a [2];
FF antagonizes AML1a-induced alteration of 32Dcl3 murine myeloid cells to granulocyte colony-stimulating factor [2];
FF ERK-dependent phosphorylation does not change DNA binding affinity, but can potentiate trans-activation ability [4];
FF reduction activates DNA-binding [5];
XX
IN T10677 FOXO3a; Mammalia.
IN T27869 MOZ; human, Homo sapiens.
IN T01427 p300; human, Homo sapiens.
IN T21984 p300; human, Homo sapiens.
IN T23091 p300; Mammalia.
IN T23072 PEBP2beta; Mammalia.
IN T14622 sin3a; human, Homo sapiens.
XX
MX M00271 V$AML1_01.
MX M01658 V$AML1_Q4.
MX M07242 V$AML1_Q4_01.
MX M08865 V$AML1_Q4_02.
MX M02084 V$AML1_Q5.
MX M08866 V$AML_Q4.
MX M00769 V$AML_Q6.
MX M00984 V$PEBP_Q6.
XX
BS R18667.
BS R04442.
BS R60225.
BS R37197.
BS R37200.
BS R23683.
BS R18666.
BS R73985.
XX
DR TRANSPATH: MO000026253.
DR TRANSCOMPEL: C00175.
DR TRANSCOMPEL: C00224.
DR TRANSCOMPEL: C00229.
DR TRANSCOMPEL: C00230.
DR EMBL: D43968;
DR EMBL: L34598;
DR UniProtKB: Q01196-1;
XX
RN [1]; RE0006068.
RX PUBMED: 7651838.
RA Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soeda E., Ohki M.
RT Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia
RL Nucleic Acids Res. 23:2762-2769 (1995).
RN [2]; RE0006069.
RX PUBMED: 7530657.
RA Tanaka T., Tanaka K., Ogawa S., Kurokawa M., Mitani K., Nishida J., Shibata Y., Yazaki Y., Hirai H.
RT An acute myleoid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms
RL EMBO J. 14:341-350 (1995).
RN [3]; RE0006082.
RX PUBMED: 8654962.
RA Ahn M., Bae S., Maruyama M., Ito Y.
RT Comparison of the human genomic structure of the Runt domain-encoding PEBP2/CBFalpha gene family
RL Gene 168:279-280 (1996).
RN [4]; RE0006087.
RX PUBMED: 8668214.
RA Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., Hirai H.
RT The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactvation ability
RL Mol. Cell. Biol. 16:3967-3979 (1996).
RN [5]; RE0006206.
RX PUBMED: 8663420.
RA Kurokawa M., Tanaka T., Tanaka K., Hirano N., Ogawa S., Mitani K., Yazaki Y., Hirai H.
RT Aconserved cystein residue in the runt homology domain of AML1 is required for the DNA binding ability and the transforming activity on fibroblasts
RL J. Biol. Chem. 271:16870-16876 (1996).
RN [6]; RE0006488.
RX PUBMED: 9192636.
RA Zeng C., van Wijnen A. J., Stein J. L., Meyers S., Sun W., Shopland L., Lawrence J. B., Penman S., Lian J.B., Stein G. S., Hiebert S. W.
RT Identification of a nuclear matrix targeting signal in leukemia and bone-related AML/CBF-alpha transcription factors
RL Proc. Natl. Acad. Sci. USA 94:6746-6751 (1997).
RN [7]; RE0039427.
RX PUBMED: 9606182.
RA Kitabayashi I., Yokoyama A., Shimizu K., Ohki M.
RT Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation
RL EMBO J. 17:2994-3004 (1998).
RN [8]; RE0039460.
RX PUBMED: 10617663.
RA Lutterbach B., Westendorf J. J., Linggi B., Isaac S., Seto E., Hiebert S. W.
RT A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia
RL J. Biol. Chem. 275:651-6 (2000).
RN [9]; RE0049455.
RX PUBMED: 16917507.
RA Aikawa Y., Nguyen L. A., Isono K., Takakura N., Tagata Y., Schmitz M. L., Koseki H., Kitabayashi I.
RT Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation.
RL EMBO J. 25:3955-3965 (2006).
RN [10]; RE0035612.
RX PUBMED: 16338472.
RA Schaubach B. M., Wen H. Y., Kellems R. E.
RT Regulation of Murine Ada Gene Expression in the Placenta by Transcription Factor RUNX1.
RL Placenta 27:269-277 (2006).
XX
//
XX
SF very similar to PEBP2alphaB from mouse [1];
SF product of AML1 gene as the result of differential usage of polyadenylation signals [1];
SF related products are AML1a T02256 and AML1c T02246 [1];
SF harbors free SH-groups [5];
XX
CN brain [1], heart [1] [1].
XX
FF nuclear extracts from HTR-8/SV*NEO, JEG-3 and embryonic day 14 placentas were all positive for Runx1b (AML1b), the active form of RUNX [10];
FF expression level of AML1b vary between different tissues [1];
FF related to t(8,21) acute myeloid leukemia;
FF binds to PEBP2 site with lower affinity than AML1a [2];
FF AML1b-trans-activation is negatively regulated by AML1a [2];
FF antagonizes AML1a-induced alteration of 32Dcl3 murine myeloid cells to granulocyte colony-stimulating factor [2];
FF ERK-dependent phosphorylation does not change DNA binding affinity, but can potentiate trans-activation ability [4];
FF reduction activates DNA-binding [5];
XX
IN T10677 FOXO3a; Mammalia.
IN T27869 MOZ; human, Homo sapiens.
IN T01427 p300; human, Homo sapiens.
IN T21984 p300; human, Homo sapiens.
IN T23091 p300; Mammalia.
IN T23072 PEBP2beta; Mammalia.
IN T14622 sin3a; human, Homo sapiens.
XX
MX M00271 V$AML1_01.
MX M01658 V$AML1_Q4.
MX M07242 V$AML1_Q4_01.
MX M08865 V$AML1_Q4_02.
MX M02084 V$AML1_Q5.
MX M08866 V$AML_Q4.
MX M00769 V$AML_Q6.
MX M00984 V$PEBP_Q6.
XX
BS R18667.
BS R04442.
BS R60225.
BS R37197.
BS R37200.
BS R23683.
BS R18666.
BS R73985.
XX
DR TRANSPATH: MO000026253.
DR TRANSCOMPEL: C00175.
DR TRANSCOMPEL: C00224.
DR TRANSCOMPEL: C00229.
DR TRANSCOMPEL: C00230.
DR EMBL: D43968;
DR EMBL: L34598;
DR UniProtKB: Q01196-1;
XX
RN [1]; RE0006068.
RX PUBMED: 7651838.
RA Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soeda E., Ohki M.
RT Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia
RL Nucleic Acids Res. 23:2762-2769 (1995).
RN [2]; RE0006069.
RX PUBMED: 7530657.
RA Tanaka T., Tanaka K., Ogawa S., Kurokawa M., Mitani K., Nishida J., Shibata Y., Yazaki Y., Hirai H.
RT An acute myleoid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms
RL EMBO J. 14:341-350 (1995).
RN [3]; RE0006082.
RX PUBMED: 8654962.
RA Ahn M., Bae S., Maruyama M., Ito Y.
RT Comparison of the human genomic structure of the Runt domain-encoding PEBP2/CBFalpha gene family
RL Gene 168:279-280 (1996).
RN [4]; RE0006087.
RX PUBMED: 8668214.
RA Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., Hirai H.
RT The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactvation ability
RL Mol. Cell. Biol. 16:3967-3979 (1996).
RN [5]; RE0006206.
RX PUBMED: 8663420.
RA Kurokawa M., Tanaka T., Tanaka K., Hirano N., Ogawa S., Mitani K., Yazaki Y., Hirai H.
RT Aconserved cystein residue in the runt homology domain of AML1 is required for the DNA binding ability and the transforming activity on fibroblasts
RL J. Biol. Chem. 271:16870-16876 (1996).
RN [6]; RE0006488.
RX PUBMED: 9192636.
RA Zeng C., van Wijnen A. J., Stein J. L., Meyers S., Sun W., Shopland L., Lawrence J. B., Penman S., Lian J.B., Stein G. S., Hiebert S. W.
RT Identification of a nuclear matrix targeting signal in leukemia and bone-related AML/CBF-alpha transcription factors
RL Proc. Natl. Acad. Sci. USA 94:6746-6751 (1997).
RN [7]; RE0039427.
RX PUBMED: 9606182.
RA Kitabayashi I., Yokoyama A., Shimizu K., Ohki M.
RT Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation
RL EMBO J. 17:2994-3004 (1998).
RN [8]; RE0039460.
RX PUBMED: 10617663.
RA Lutterbach B., Westendorf J. J., Linggi B., Isaac S., Seto E., Hiebert S. W.
RT A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia
RL J. Biol. Chem. 275:651-6 (2000).
RN [9]; RE0049455.
RX PUBMED: 16917507.
RA Aikawa Y., Nguyen L. A., Isono K., Takakura N., Tagata Y., Schmitz M. L., Koseki H., Kitabayashi I.
RT Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation.
RL EMBO J. 25:3955-3965 (2006).
RN [10]; RE0035612.
RX PUBMED: 16338472.
RA Schaubach B. M., Wen H. Y., Kellems R. E.
RT Regulation of Murine Ada Gene Expression in the Placenta by Transcription Factor RUNX1.
RL Placenta 27:269-277 (2006).
XX
//


PF00853; Runt domain. FT 50 178
PS51062; RUNT. FT 66 438
PF00478; IMP dehydrogenase / GMP reductase domain. FT 72 72
C is important for DNA-binding and biological activity [5]. FT 249 249
major extracellular signal-regulated kinase (ERK)-dependent phosphorylation site [4]. FT 266 266
minor ERK-dependent phosphorylation site [4]. FT 324 354
putative nuclear matrix targeting signal [6]. FT 392 392
S replaced by F [3].
XX SF very similar to PEBP2alphaB from mouse [1]; SF product of AML1 gene as the result of differential usage of polyadenylation signals [1]; SF related products are AML1a T02256 and AML1c T02246 [1]; SF harbors free SH-groups [5]; XX CN brain [1], heart [1] [1]. XX FF nuclear extracts from HTR-8/SV*NEO, JEG-3 and embryonic day 14 placentas were all positive for Runx1b (AML1b), the active form of RUNX [10]; FF expression level of AML1b vary between different tissues [1]; FF related to t(8,21) acute myeloid leukemia; FF binds to PEBP2 site with lower affinity than AML1a [2]; FF AML1b-trans-activation is negatively regulated by AML1a [2]; FF antagonizes AML1a-induced alteration of 32Dcl3 murine myeloid cells to granulocyte colony-stimulating factor [2]; FF ERK-dependent phosphorylation does not change DNA binding affinity, but can potentiate trans-activation ability [4]; FF reduction activates DNA-binding [5]; XX IN T10677 FOXO3a; Mammalia. IN T27869 MOZ; human, Homo sapiens. IN T01427 p300; human, Homo sapiens. IN T21984 p300; human, Homo sapiens. IN T23091 p300; Mammalia. IN T23072 PEBP2beta; Mammalia. IN T14622 sin3a; human, Homo sapiens. XX MX M00271 V$AML1_01. MX M01658 V$AML1_Q4. MX M07242 V$AML1_Q4_01. MX M08865 V$AML1_Q4_02. MX M02084 V$AML1_Q5. MX M08866 V$AML_Q4. MX M00769 V$AML_Q6. MX M00984 V$PEBP_Q6. XX BS R18667. BS R04442. BS R60225. BS R37197. BS R37200. BS R23683. BS R18666. BS R73985. XX DR TRANSPATH: MO000026253. DR TRANSCOMPEL: C00175. DR TRANSCOMPEL: C00224. DR TRANSCOMPEL: C00229. DR TRANSCOMPEL: C00230. DR EMBL: D43968; DR EMBL: L34598; DR UniProtKB: Q01196-1; XX RN [1]; RE0006068. RX PUBMED: 7651838. RA Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soeda E., Ohki M. RT Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia RL Nucleic Acids Res. 23:2762-2769 (1995). RN [2]; RE0006069. RX PUBMED: 7530657. RA Tanaka T., Tanaka K., Ogawa S., Kurokawa M., Mitani K., Nishida J., Shibata Y., Yazaki Y., Hirai H. RT An acute myleoid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms RL EMBO J. 14:341-350 (1995). RN [3]; RE0006082. RX PUBMED: 8654962. RA Ahn M., Bae S., Maruyama M., Ito Y. RT Comparison of the human genomic structure of the Runt domain-encoding PEBP2/CBFalpha gene family RL Gene 168:279-280 (1996). RN [4]; RE0006087. RX PUBMED: 8668214. RA Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., Kadowaki T., Hirai H. RT The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactvation ability RL Mol. Cell. Biol. 16:3967-3979 (1996). RN [5]; RE0006206. RX PUBMED: 8663420. RA Kurokawa M., Tanaka T., Tanaka K., Hirano N., Ogawa S., Mitani K., Yazaki Y., Hirai H. RT Aconserved cystein residue in the runt homology domain of AML1 is required for the DNA binding ability and the transforming activity on fibroblasts RL J. Biol. Chem. 271:16870-16876 (1996). RN [6]; RE0006488. RX PUBMED: 9192636. RA Zeng C., van Wijnen A. J., Stein J. L., Meyers S., Sun W., Shopland L., Lawrence J. B., Penman S., Lian J.B., Stein G. S., Hiebert S. W. RT Identification of a nuclear matrix targeting signal in leukemia and bone-related AML/CBF-alpha transcription factors RL Proc. Natl. Acad. Sci. USA 94:6746-6751 (1997). RN [7]; RE0039427. RX PUBMED: 9606182. RA Kitabayashi I., Yokoyama A., Shimizu K., Ohki M. RT Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation RL EMBO J. 17:2994-3004 (1998). RN [8]; RE0039460. RX PUBMED: 10617663. RA Lutterbach B., Westendorf J. J., Linggi B., Isaac S., Seto E., Hiebert S. W. RT A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia RL J. Biol. Chem. 275:651-6 (2000). RN [9]; RE0049455. RX PUBMED: 16917507. RA Aikawa Y., Nguyen L. A., Isono K., Takakura N., Tagata Y., Schmitz M. L., Koseki H., Kitabayashi I. RT Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. RL EMBO J. 25:3955-3965 (2006). RN [10]; RE0035612. RX PUBMED: 16338472. RA Schaubach B. M., Wen H. Y., Kellems R. E. RT Regulation of Murine Ada Gene Expression in the Placenta by Transcription Factor RUNX1. RL Placenta 27:269-277 (2006). XX //