
AC T01567
XX
ID T01567
XX
DT 19.09.1995 (created); ewi.
DT 12.01.2010 (updated); jig.
CO Copyright (C), QIAGEN.
XX
FA Max-isoform1
XX
SY Max2; Myn; p22.
XX
OS human, Homo sapiens
OC eukaryota; animalia; metazoa; chordata; vertebrata; tetrapoda; mammalia; eutheria; primates
XX
GE G003958 MAX; HGNC: Max.
XX
CL C0012; bHLH-ZIP; 1.2.6.5.5.1.
XX
SZ 160 AA; 18.3 kDa (cDNA) (calc.), 22 kDa (SDS) [8]
XX
SQ MSDNDDIEVESDEEQPRFQSAADKRAHHNALERKRRDHIKDSFHSLRDSVPSLQGEKASR
SQ AQILDKATEYIQYMRRKNHTHQQDIDDLKRQNALLEQQVRALEKARSSAQLQTNYPSSDN
SQ SLYTNAKGSTISAFDGGSDSSSESEPEEPQSRKKLRMEAS
XX
SC Swiss-Prot#P61244-1
XX
FT 2 2  Ser-phosphorylation by CKII [16].
FT 11 11
Ser-phosphorylation by CKII [16].
FT 11 11  Ser-phosphorylation by CKII [16].
FT 24 75
Ser-phosphorylation by CKII [16].
FT 24 75  PF00010; Helix-loop-helix DNA-binding domain.
FT 24 75
PF00010; Helix-loop-helix DNA-binding domain.
FT 24 75  PS50888; HLH.
FT 29 80
PS50888; HLH.
FT 29 80  SM00353; finulus.
FT 99 160
SM00353; finulus.
FT 99 160  replaced in delta Max by GESES [7].
FT 149 156
replaced in delta Max by GESES [7].
FT 149 156  nuclear localization signal (NLS) [2].
nuclear localization signal (NLS) [2].
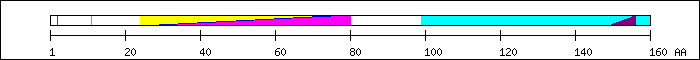 XX
SF several splice variants T00489, T01567 detected;
SF Max homodimers and Myc/Max heterodimers exhibit different preferences for the trinucleotides flanking the core sequence (CACGTG) [5];
SF Max1 and Max-isoform1 also differ slightly in their DNA-binding specificity [5] [20];
SF heterodimers with either c-Myc or Mad exert higher DNA-binding affinity than Max homodimers [17];
SF heterodimerization requires the leucine zipper, but not homodimerization [13] [2];
SF DNA-binding by Max or Myc/Max induces bending towards the minor groove by 74-82 deg, but possibly in opposite orientations [6] [12];
SF belongs to group B HLH-proteins that bind to CACGTG motif in DNA which is due to certain residues in DNA-binding domain (His at pos. 28, Glu at pos. 32, Arg at pos. 36) [23] [24];
XX
FF enhances c-Myc DNA-binding;
FF in high concentrations: homodimers suppress c-Myc activities by competition [7] [9] [2];
FF after CKII phosphorylation, increases in on- and off-rates of DNA-binding of homo- and heterodimers have been found [16];
FF preferentially complexed with Mad instead of c-Myc upon myeloid differentiation [19];
XX
IN T00140 c-Myc-isoform1; human, Homo sapiens.
IN T01565 Mad1; human, Homo sapiens.
IN T02390 Mad1; mouse, Mus musculus.
IN T01567 Max-isoform1; human, Homo sapiens.
IN T01566 Max-isoform3; human, Homo sapiens.
IN T01564 Mxi1; human, Homo sapiens.
IN T01445 N-Myc; mouse, Mus musculus.
IN T02379 N-Myc; human, Homo sapiens.
IN T09033 TEF-1; human, Homo sapiens.
XX
MX M01034 V$EBOX_Q6_01.
MX M00119 V$MAX_01.
MX M00118 V$MYCMAX_01.
MX M00123 V$MYCMAX_02.
MX M00615 V$MYCMAX_03.
MX M00322 V$MYCMAX_B.
MX M00799 V$MYC_Q2.
XX
DR TRANSPATH: MO000019559.
DR EMBL: M64240; HSMAX.
DR UniProtKB: P61244-1;
XX
RN [1]; RE0000477.
RX PUBMED: 1935896.
RA Wenzel A., Cziepluch C., Hamann U., Schuermann J., Schwab M.
RT The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells
RL EMBO J. 10:3703-3712 (1991).
RN [2]; RE0000704.
RX PUBMED: 1730412.
RA Kato G. J., Lee W. M. F., Chen L., Dang C. V.
RT Max: functional domains and interaction with c-Myc
RL Genes Dev. 6:81-92 (1992).
RN [3]; RE0000705.
RX PUBMED: 1737614.
RA Berberich S. J., Cole M. D.
RT Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers
RL Genes Dev. 6:166-176 (1992).
RN [4]; RE0002653.
RX PUBMED: 2006410.
RA Blackwood E. M., Eisenman R. N.
RT Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc
RL Science 251:1211-1217 (1991).
RN [5]; RE0002937.
RX PUBMED: 8265351.
RA Solomon D. L. C., Amati B., Land H.
RT Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers
RL Nucleic Acids Res. 21:5372-5376 (1993).
RN [6]; RE0003338.
RX PUBMED: 1465398.
RA Fisher D. E., Parent L. A., Sharp P. A.
RT Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove
RL Proc. Natl. Acad. Sci. USA 89:11779-11783 (1992).
RN [7]; RE0003350.
RX PUBMED: 1566084.
RA Maekelae T. P., Koskinen P. J., Vaestrik I., Alitalo K.
RT Alternative forms of Max as enhancers or suppressors of Myc-Ras cotransformation
RL Science 256:373-377 (1992).
RN [8]; RE0003353.
RX PUBMED: 1730411.
RA Blackwood E. M., Luescher B., Eisenman R. N.
RT Myc and Max associate in vivo
RL Genes Dev. 6:71-80 (1992).
RN [9]; RE0003361.
RX PUBMED: 8425220.
RA Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H.
RT Oncogenic activity of the c-Myc protein requires dimerization with Max
RL Cell 72:233-245 (1993).
RN [10]; RE0003387.
RX PUBMED: 1630816.
RA Min S., Taparowsky E. J.
RT v-Myc, but not Max, possesses domains that function in both transcription activation and cellular transformation
RL Oncogene 7:1531-1540 (1992).
RN [11]; RE0003395.
RX PUBMED: 1459463.
RA Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B.
RT Biphasic effect of Max and Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions
RL Genes Dev. 6:2429-2439 (1992).
RN [12]; RE0003396.
RX PUBMED: 1323849.
RA Wechsler D. S., Dang C. V.
RT Opposite directions of DNA bending by c-Myc and Max
RL Proc. Natl. Acad. Sci. USA 89:7635-7639 (1992).
RN [13]; RE0003397.
RX PUBMED: 1408152.
RA Reddy C. D., Dasgupta P., Saikumar P., Dudek H., Rauscher III F. J., Reddy E. P.
RT Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation
RL Oncogene 7:2085-2092 (1992).
RN [14]; RE0003398.
RX PUBMED: 1557420.
RA Wagner A. J., Le Beau M. M., Diaz M. O., Hay N.
RT Expression, regulation, and chromosomal localization of the Max gene
RL Proc. Natl. Acad. Sci. USA 89:3111-3115 (1992).
RN [15]; RE0003399.
RX PUBMED: 8262050.
RA Fisher F., Crouch D. H., Jayaraman P. S., Clark W., Gillespie D. A. F., Goding C. R.
RT Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo
RL EMBO J. 12:5075-5082 (1993).
RN [16]; RE0003400.
RX PUBMED: 8247525.
RA Bousset K., Henriksson M., Luescher-Firzlaff J. M., Litchfield D. W., Luescher B.
RT Identification of casein kinase II phosphorylation sites in Max: effects on DNA-binding kinetics of Max homo- and Myc/Max heterodimers
RL Oncogene 8:3211-3220 (1993).
RN [17]; RE0003401.
RX PUBMED: 8425218.
RA Ayer D. E., Kretzner L., Eisenman R. N.
RT Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity
RL Cell 72:211-222 (1993).
RN [18]; RE0003402.
RX PUBMED: 8425219.
RA Zervos A. S., Gyuris J., Brent R.
RT Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites
RL Cell 72:223-232 (1993).
RN [19]; RE0003403.
RX PUBMED: 8224841.
RA Ayer D. E., Eisenman R. N.
RT A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation
RL Genes Dev. 7:2110-2119 (1993).
RN [20]; RE0003404.
RX PUBMED: 8430110.
RA Prochownik E. V., Antwerp M. E.
RT Differential patterns of DNA binding by myc and max proteins
RL Proc. Natl. Acad. Sci. USA 90:960-964 (1993).
RN [21]; RE0014494.
RX PUBMED: 2040011.
RA Cole M. D.
RT Myc meets its Max
RL Cell 65:715-716 (1991).
RN [22]; RE0014787.
RX PUBMED: 8290277.
RA Sollenberger K. G., Kao T. L., Taparowsky E. J.
RT Structural analysis of the chicken max gene
RL Oncogene 9:661-664 (1994).
RN [23]; RE0017805.
RX PUBMED: 9860302.
RA Meng X., Lu X., Li Z., Green E. D., Massa H., Trask B. J., Morris C. A., Keating M. T.
RT Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes
RL Hum. Genet. 103:590-599 (1998).
RN [24]; RE0017817.
RX PUBMED: 11073985.
RA Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E.
RT MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network
RL Mol. Cell. Biol. 20:8845-8854 (2000).
RN [25]; RE0047848.
RX PUBMED: 10931933.
RA Gupta M. P., Kogut P., Gupta M.
RT Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability.
RL Nucleic Acids Res. 28:3168-3177 (2000).
XX
//
XX
SF several splice variants T00489, T01567 detected;
SF Max homodimers and Myc/Max heterodimers exhibit different preferences for the trinucleotides flanking the core sequence (CACGTG) [5];
SF Max1 and Max-isoform1 also differ slightly in their DNA-binding specificity [5] [20];
SF heterodimers with either c-Myc or Mad exert higher DNA-binding affinity than Max homodimers [17];
SF heterodimerization requires the leucine zipper, but not homodimerization [13] [2];
SF DNA-binding by Max or Myc/Max induces bending towards the minor groove by 74-82 deg, but possibly in opposite orientations [6] [12];
SF belongs to group B HLH-proteins that bind to CACGTG motif in DNA which is due to certain residues in DNA-binding domain (His at pos. 28, Glu at pos. 32, Arg at pos. 36) [23] [24];
XX
FF enhances c-Myc DNA-binding;
FF in high concentrations: homodimers suppress c-Myc activities by competition [7] [9] [2];
FF after CKII phosphorylation, increases in on- and off-rates of DNA-binding of homo- and heterodimers have been found [16];
FF preferentially complexed with Mad instead of c-Myc upon myeloid differentiation [19];
XX
IN T00140 c-Myc-isoform1; human, Homo sapiens.
IN T01565 Mad1; human, Homo sapiens.
IN T02390 Mad1; mouse, Mus musculus.
IN T01567 Max-isoform1; human, Homo sapiens.
IN T01566 Max-isoform3; human, Homo sapiens.
IN T01564 Mxi1; human, Homo sapiens.
IN T01445 N-Myc; mouse, Mus musculus.
IN T02379 N-Myc; human, Homo sapiens.
IN T09033 TEF-1; human, Homo sapiens.
XX
MX M01034 V$EBOX_Q6_01.
MX M00119 V$MAX_01.
MX M00118 V$MYCMAX_01.
MX M00123 V$MYCMAX_02.
MX M00615 V$MYCMAX_03.
MX M00322 V$MYCMAX_B.
MX M00799 V$MYC_Q2.
XX
DR TRANSPATH: MO000019559.
DR EMBL: M64240; HSMAX.
DR UniProtKB: P61244-1;
XX
RN [1]; RE0000477.
RX PUBMED: 1935896.
RA Wenzel A., Cziepluch C., Hamann U., Schuermann J., Schwab M.
RT The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells
RL EMBO J. 10:3703-3712 (1991).
RN [2]; RE0000704.
RX PUBMED: 1730412.
RA Kato G. J., Lee W. M. F., Chen L., Dang C. V.
RT Max: functional domains and interaction with c-Myc
RL Genes Dev. 6:81-92 (1992).
RN [3]; RE0000705.
RX PUBMED: 1737614.
RA Berberich S. J., Cole M. D.
RT Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers
RL Genes Dev. 6:166-176 (1992).
RN [4]; RE0002653.
RX PUBMED: 2006410.
RA Blackwood E. M., Eisenman R. N.
RT Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc
RL Science 251:1211-1217 (1991).
RN [5]; RE0002937.
RX PUBMED: 8265351.
RA Solomon D. L. C., Amati B., Land H.
RT Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers
RL Nucleic Acids Res. 21:5372-5376 (1993).
RN [6]; RE0003338.
RX PUBMED: 1465398.
RA Fisher D. E., Parent L. A., Sharp P. A.
RT Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove
RL Proc. Natl. Acad. Sci. USA 89:11779-11783 (1992).
RN [7]; RE0003350.
RX PUBMED: 1566084.
RA Maekelae T. P., Koskinen P. J., Vaestrik I., Alitalo K.
RT Alternative forms of Max as enhancers or suppressors of Myc-Ras cotransformation
RL Science 256:373-377 (1992).
RN [8]; RE0003353.
RX PUBMED: 1730411.
RA Blackwood E. M., Luescher B., Eisenman R. N.
RT Myc and Max associate in vivo
RL Genes Dev. 6:71-80 (1992).
RN [9]; RE0003361.
RX PUBMED: 8425220.
RA Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H.
RT Oncogenic activity of the c-Myc protein requires dimerization with Max
RL Cell 72:233-245 (1993).
RN [10]; RE0003387.
RX PUBMED: 1630816.
RA Min S., Taparowsky E. J.
RT v-Myc, but not Max, possesses domains that function in both transcription activation and cellular transformation
RL Oncogene 7:1531-1540 (1992).
RN [11]; RE0003395.
RX PUBMED: 1459463.
RA Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B.
RT Biphasic effect of Max and Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions
RL Genes Dev. 6:2429-2439 (1992).
RN [12]; RE0003396.
RX PUBMED: 1323849.
RA Wechsler D. S., Dang C. V.
RT Opposite directions of DNA bending by c-Myc and Max
RL Proc. Natl. Acad. Sci. USA 89:7635-7639 (1992).
RN [13]; RE0003397.
RX PUBMED: 1408152.
RA Reddy C. D., Dasgupta P., Saikumar P., Dudek H., Rauscher III F. J., Reddy E. P.
RT Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation
RL Oncogene 7:2085-2092 (1992).
RN [14]; RE0003398.
RX PUBMED: 1557420.
RA Wagner A. J., Le Beau M. M., Diaz M. O., Hay N.
RT Expression, regulation, and chromosomal localization of the Max gene
RL Proc. Natl. Acad. Sci. USA 89:3111-3115 (1992).
RN [15]; RE0003399.
RX PUBMED: 8262050.
RA Fisher F., Crouch D. H., Jayaraman P. S., Clark W., Gillespie D. A. F., Goding C. R.
RT Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo
RL EMBO J. 12:5075-5082 (1993).
RN [16]; RE0003400.
RX PUBMED: 8247525.
RA Bousset K., Henriksson M., Luescher-Firzlaff J. M., Litchfield D. W., Luescher B.
RT Identification of casein kinase II phosphorylation sites in Max: effects on DNA-binding kinetics of Max homo- and Myc/Max heterodimers
RL Oncogene 8:3211-3220 (1993).
RN [17]; RE0003401.
RX PUBMED: 8425218.
RA Ayer D. E., Kretzner L., Eisenman R. N.
RT Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity
RL Cell 72:211-222 (1993).
RN [18]; RE0003402.
RX PUBMED: 8425219.
RA Zervos A. S., Gyuris J., Brent R.
RT Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites
RL Cell 72:223-232 (1993).
RN [19]; RE0003403.
RX PUBMED: 8224841.
RA Ayer D. E., Eisenman R. N.
RT A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation
RL Genes Dev. 7:2110-2119 (1993).
RN [20]; RE0003404.
RX PUBMED: 8430110.
RA Prochownik E. V., Antwerp M. E.
RT Differential patterns of DNA binding by myc and max proteins
RL Proc. Natl. Acad. Sci. USA 90:960-964 (1993).
RN [21]; RE0014494.
RX PUBMED: 2040011.
RA Cole M. D.
RT Myc meets its Max
RL Cell 65:715-716 (1991).
RN [22]; RE0014787.
RX PUBMED: 8290277.
RA Sollenberger K. G., Kao T. L., Taparowsky E. J.
RT Structural analysis of the chicken max gene
RL Oncogene 9:661-664 (1994).
RN [23]; RE0017805.
RX PUBMED: 9860302.
RA Meng X., Lu X., Li Z., Green E. D., Massa H., Trask B. J., Morris C. A., Keating M. T.
RT Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes
RL Hum. Genet. 103:590-599 (1998).
RN [24]; RE0017817.
RX PUBMED: 11073985.
RA Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E.
RT MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network
RL Mol. Cell. Biol. 20:8845-8854 (2000).
RN [25]; RE0047848.
RX PUBMED: 10931933.
RA Gupta M. P., Kogut P., Gupta M.
RT Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability.
RL Nucleic Acids Res. 28:3168-3177 (2000).
XX
//


Ser-phosphorylation by CKII [16]. FT 11 11
Ser-phosphorylation by CKII [16]. FT 24 75
PF00010; Helix-loop-helix DNA-binding domain. FT 24 75
PS50888; HLH. FT 29 80
SM00353; finulus. FT 99 160
replaced in delta Max by GESES [7]. FT 149 156
nuclear localization signal (NLS) [2].
XX SF several splice variants T00489, T01567 detected; SF Max homodimers and Myc/Max heterodimers exhibit different preferences for the trinucleotides flanking the core sequence (CACGTG) [5]; SF Max1 and Max-isoform1 also differ slightly in their DNA-binding specificity [5] [20]; SF heterodimers with either c-Myc or Mad exert higher DNA-binding affinity than Max homodimers [17]; SF heterodimerization requires the leucine zipper, but not homodimerization [13] [2]; SF DNA-binding by Max or Myc/Max induces bending towards the minor groove by 74-82 deg, but possibly in opposite orientations [6] [12]; SF belongs to group B HLH-proteins that bind to CACGTG motif in DNA which is due to certain residues in DNA-binding domain (His at pos. 28, Glu at pos. 32, Arg at pos. 36) [23] [24]; XX FF enhances c-Myc DNA-binding; FF in high concentrations: homodimers suppress c-Myc activities by competition [7] [9] [2]; FF after CKII phosphorylation, increases in on- and off-rates of DNA-binding of homo- and heterodimers have been found [16]; FF preferentially complexed with Mad instead of c-Myc upon myeloid differentiation [19]; XX IN T00140 c-Myc-isoform1; human, Homo sapiens. IN T01565 Mad1; human, Homo sapiens. IN T02390 Mad1; mouse, Mus musculus. IN T01567 Max-isoform1; human, Homo sapiens. IN T01566 Max-isoform3; human, Homo sapiens. IN T01564 Mxi1; human, Homo sapiens. IN T01445 N-Myc; mouse, Mus musculus. IN T02379 N-Myc; human, Homo sapiens. IN T09033 TEF-1; human, Homo sapiens. XX MX M01034 V$EBOX_Q6_01. MX M00119 V$MAX_01. MX M00118 V$MYCMAX_01. MX M00123 V$MYCMAX_02. MX M00615 V$MYCMAX_03. MX M00322 V$MYCMAX_B. MX M00799 V$MYC_Q2. XX DR TRANSPATH: MO000019559. DR EMBL: M64240; HSMAX. DR UniProtKB: P61244-1; XX RN [1]; RE0000477. RX PUBMED: 1935896. RA Wenzel A., Cziepluch C., Hamann U., Schuermann J., Schwab M. RT The N-Myc oncoprotein is associated in vivo with the phosphoprotein Max(p20/22) in human neuroblastoma cells RL EMBO J. 10:3703-3712 (1991). RN [2]; RE0000704. RX PUBMED: 1730412. RA Kato G. J., Lee W. M. F., Chen L., Dang C. V. RT Max: functional domains and interaction with c-Myc RL Genes Dev. 6:81-92 (1992). RN [3]; RE0000705. RX PUBMED: 1737614. RA Berberich S. J., Cole M. D. RT Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers RL Genes Dev. 6:166-176 (1992). RN [4]; RE0002653. RX PUBMED: 2006410. RA Blackwood E. M., Eisenman R. N. RT Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc RL Science 251:1211-1217 (1991). RN [5]; RE0002937. RX PUBMED: 8265351. RA Solomon D. L. C., Amati B., Land H. RT Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers RL Nucleic Acids Res. 21:5372-5376 (1993). RN [6]; RE0003338. RX PUBMED: 1465398. RA Fisher D. E., Parent L. A., Sharp P. A. RT Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove RL Proc. Natl. Acad. Sci. USA 89:11779-11783 (1992). RN [7]; RE0003350. RX PUBMED: 1566084. RA Maekelae T. P., Koskinen P. J., Vaestrik I., Alitalo K. RT Alternative forms of Max as enhancers or suppressors of Myc-Ras cotransformation RL Science 256:373-377 (1992). RN [8]; RE0003353. RX PUBMED: 1730411. RA Blackwood E. M., Luescher B., Eisenman R. N. RT Myc and Max associate in vivo RL Genes Dev. 6:71-80 (1992). RN [9]; RE0003361. RX PUBMED: 8425220. RA Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H. RT Oncogenic activity of the c-Myc protein requires dimerization with Max RL Cell 72:233-245 (1993). RN [10]; RE0003387. RX PUBMED: 1630816. RA Min S., Taparowsky E. J. RT v-Myc, but not Max, possesses domains that function in both transcription activation and cellular transformation RL Oncogene 7:1531-1540 (1992). RN [11]; RE0003395. RX PUBMED: 1459463. RA Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B. RT Biphasic effect of Max and Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions RL Genes Dev. 6:2429-2439 (1992). RN [12]; RE0003396. RX PUBMED: 1323849. RA Wechsler D. S., Dang C. V. RT Opposite directions of DNA bending by c-Myc and Max RL Proc. Natl. Acad. Sci. USA 89:7635-7639 (1992). RN [13]; RE0003397. RX PUBMED: 1408152. RA Reddy C. D., Dasgupta P., Saikumar P., Dudek H., Rauscher III F. J., Reddy E. P. RT Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation RL Oncogene 7:2085-2092 (1992). RN [14]; RE0003398. RX PUBMED: 1557420. RA Wagner A. J., Le Beau M. M., Diaz M. O., Hay N. RT Expression, regulation, and chromosomal localization of the Max gene RL Proc. Natl. Acad. Sci. USA 89:3111-3115 (1992). RN [15]; RE0003399. RX PUBMED: 8262050. RA Fisher F., Crouch D. H., Jayaraman P. S., Clark W., Gillespie D. A. F., Goding C. R. RT Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo RL EMBO J. 12:5075-5082 (1993). RN [16]; RE0003400. RX PUBMED: 8247525. RA Bousset K., Henriksson M., Luescher-Firzlaff J. M., Litchfield D. W., Luescher B. RT Identification of casein kinase II phosphorylation sites in Max: effects on DNA-binding kinetics of Max homo- and Myc/Max heterodimers RL Oncogene 8:3211-3220 (1993). RN [17]; RE0003401. RX PUBMED: 8425218. RA Ayer D. E., Kretzner L., Eisenman R. N. RT Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity RL Cell 72:211-222 (1993). RN [18]; RE0003402. RX PUBMED: 8425219. RA Zervos A. S., Gyuris J., Brent R. RT Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites RL Cell 72:223-232 (1993). RN [19]; RE0003403. RX PUBMED: 8224841. RA Ayer D. E., Eisenman R. N. RT A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation RL Genes Dev. 7:2110-2119 (1993). RN [20]; RE0003404. RX PUBMED: 8430110. RA Prochownik E. V., Antwerp M. E. RT Differential patterns of DNA binding by myc and max proteins RL Proc. Natl. Acad. Sci. USA 90:960-964 (1993). RN [21]; RE0014494. RX PUBMED: 2040011. RA Cole M. D. RT Myc meets its Max RL Cell 65:715-716 (1991). RN [22]; RE0014787. RX PUBMED: 8290277. RA Sollenberger K. G., Kao T. L., Taparowsky E. J. RT Structural analysis of the chicken max gene RL Oncogene 9:661-664 (1994). RN [23]; RE0017805. RX PUBMED: 9860302. RA Meng X., Lu X., Li Z., Green E. D., Massa H., Trask B. J., Morris C. A., Keating M. T. RT Complete physical map of the common deletion region in Williams syndrome and identification and characterization of three novel genes RL Hum. Genet. 103:590-599 (1998). RN [24]; RE0017817. RX PUBMED: 11073985. RA Billin A. N., Eilers A. L., Coulter K. L., Logan J. S., Ayer D. E. RT MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network RL Mol. Cell. Biol. 20:8845-8854 (2000). RN [25]; RE0047848. RX PUBMED: 10931933. RA Gupta M. P., Kogut P., Gupta M. RT Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability. RL Nucleic Acids Res. 28:3168-3177 (2000). XX //